Visualizing replication stress in yeast models of genome instabilityDNA replication stress can promote genome instability and cancer. We test fission yeast for the effects of DNA replication stress using genetic and high-resolution microscopy methods. We have seen that interesting differences between replication mutant yeast become apparent after S-phase stress is over: the recovery phase post-instability. This suggests that the recovery phase is an important time when cells that survive might be distinguished from cells that die. We are now testing how replication instability caused by cancer chemotherapy drugs is influenced by cellular environment. Yeast is an ideal tool to determine the genetic contributions of drug-sensitivity and resistance that may promote better patient response to chemotherapeutics. Ideally, drug-surviving cells can be identified with biomarkers found during long-term, live-cell imaging. 11:30-12:30 BioSci Rm. 3110
Chromosomes and bytes of code: A computational genomics approach to identify imprinted genes on the human X chromosomeHumans and other mammals have two sets of chromosomes: one set that is maternally inherited; the other, paternally inherited. While most genes are evenly expressed, there are cases where one copy of the gene is favoured over the other. We typically think of this happening as one trait or allele being ‘dominant’ over the other. In the case of genomic imprinting, however, gene expression is determined based on parent of origin, rather than gene sequence. This results in asymmetric expression of alleles for traits inherited maternally vs. paternally. Defects in imprinted genes are responsible for a number of diseases such as: Prader-Willi syndrome, Beckwith-Wiedemann syndrome, and Angelman syndrome. To date, there have been no imprinted genes identified on the human X chromosome. This is mainly due to the experimental limitations, as well as the random X inactivation that occurs in humans. With these restrictions in mind, how do we investigate genomic imprinting on the human X chromosome? Can computers and data mining be used to complement next-gen sequencing and traditional molecular techniques? 11:30-12:30 BioSci Rm. 3110**Note: Special date, time and place! Monday, Sept. 26 from 1:30-2:30pm in the 4th floor EEB Lounge Advancements in Western Blotting Technology: Bio-Rad's Western Workflow delivers more sensitive results, faster at a lower costBio-Rad Western Blot Imager demo after the seminar 2:30-3:30pm in the Monaghan Lab BioSci Rm. 3416
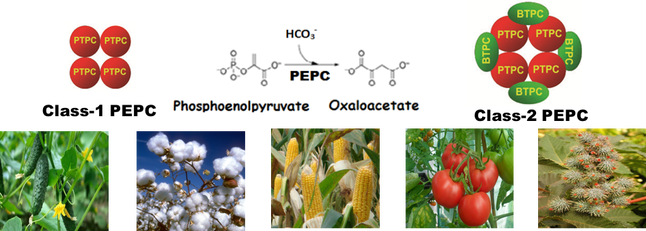
Transcriptome-Enabled Biochemistry: Unraveling the function of bacterial-type phosphoenolpyruvate carboxylase in vascular plants PEP carboxylase (PEPC) is a tightly regulated enzyme that plays essential roles in all plant cells, particularly the ‘anaplerotic’ replenishment of TCA cycle intermediates withdrawn for biosynthesis. Plant PEPC belongs to a small multigene family encoding several closely related plant-type PEPCs (PTPC) along with a distantly-related and enigmatic bacterial-type PEPC (BTPC). Previous characterization of PEPC from green microalgae and then developing castor beans led to the discovery of novel, Class-2 PEPC hetero-octameric complexes composed of tightly associated PTPC and BTPC subunits and that are largely desensitized to classic PEPC allosteric inhibitors such as malate. Mike’s bioinformatics analysis of publicly available RNA-seq and microarray transcriptomic datasets revealed two distinct patterns of tissue-specific BTPC expression. Species such as Arabidopsis, rice, maize, and poplar mainly exhibited pollen-specific BTPC expression. By contrast, BTPC transcripts were particularly abundant in non-pollen, heterotrophic ‘sink’ tissues including developing castor, cotton, soybean, and cucumber seeds, as well as the developing fruits of avocado, grape, and tomato. Follow up biochemical and co-immunoprecipitation experiments confirmed presence of ~118 kDa BTPC polypeptides that associate with co-expressed PTPC subunits to form a Class-2 PEPC complex in tomato and cucumber fruits. Overall results support the hypothesis that BTPC functions as a catalytic and regulatory subunit of Class-2 PEPCs that sustain rapid anaplerotic carboxylation of PEP while refixing respired CO2 in biosynthetically active sink tissues that accumulate abundant amounts of malate. |
Archives
February 2021
|